CFCRA Soybean Cluster: Activity 10 – A new method for precise and reproducible phenotyping of Phythophthora sojae isolates in soybean
Principal Investigator: Richard Bélanger
Research Institution: Université Laval
Objectives:
- Implementation of a new inoculation technique to assess cultivar response to P. sojae pathotypes based on zoospore infection in a miniature hydroponic system.
- Exploitation of molecular markers to validate the virulence patterns of genes present in each P. sojae isolate tested.
- Expanded analyses of all P. sojae isolates currently present in our banks to all soybean-producing regions in Canada.
Impacts:
- The phenotyping based on zoospore approach has the distinct advantage of reproducing more closely the natural infection process and, more importantly, allowing the expression of root resistance and horizontal resistance not revealed by the standard hypocotyl inoculation technique. Other advantages are the ease with which one can look at root responses in the hydroponic system, the possibility to evaluate several cultivars at once within the same system (increases reproducibility), and the possibility to test several pathotypes simultaneously. Another advantage is that any given pathotype will be consistently expressed following repeated subcultures.
- Provides precise directives for breeders to develop and exploit soybean germplasm resistant to P. sojae.
- Provides long-term recommendations to soybean breeders and growers for deployment of Rps genes in commercial soybean varieties.
- Results in reduced losses to P. sojae through use of soybean cultivars specifically resistant to pathotypes present in their growing areas.
- The technology is now used by growers and companies in Canada, USA, and Brazil to help growers select the soybean lines carrying the proper Rps genes.
Scientific Summary:
Phytophthora root rot, caused by Phytophthora sojae, is one of the most devastating diseases of soybean in Canada. The standard method of phenotyping P. sojae to identify the virulence profile, the hypocotyl assay, is hampered by several limitations.
This research activity proposed to overcome those limitations by developing the first comprehensive phenotyping of P. sojae isolates present in Canadian soybean fields based on a novel reproducible hydroponic bioassay and molecular tools.
Results:
- A novel hydroponic assay was developed to assess soybean response to sojae with better accuracy and reproducibility for both vertical (Rps genes) and horizontal resistance genes.
- A direct link was established between genomic signatures and phenotypes of sojae isolates to quickly and accurately identify virulence patterns against Rps genes.
- A molecular tool was developed to allow the simultaneous diagnosis of six avirulence genes in each sojae isolate matching the most common Rps genes (1a, 1b,1c, 1k, 3a, 6).
- Comprehensive surveys of sojae presence in soybean fields were conducted across Alberta, Saskatchewan, Manitoba, Ontario, Quebec, and Atlantic Canada in 2018, 2019, 2020, and 2021.
- The application of the molecular diagnostics tool was made available for the first time in a commercial context in 2022, to diagnose sojae pathotypes in soybean fields.
- A comprehensive map of distribution and characteristics (virulence patterns) of sojae isolates throughout soybean-producing regions in Canada was produced.
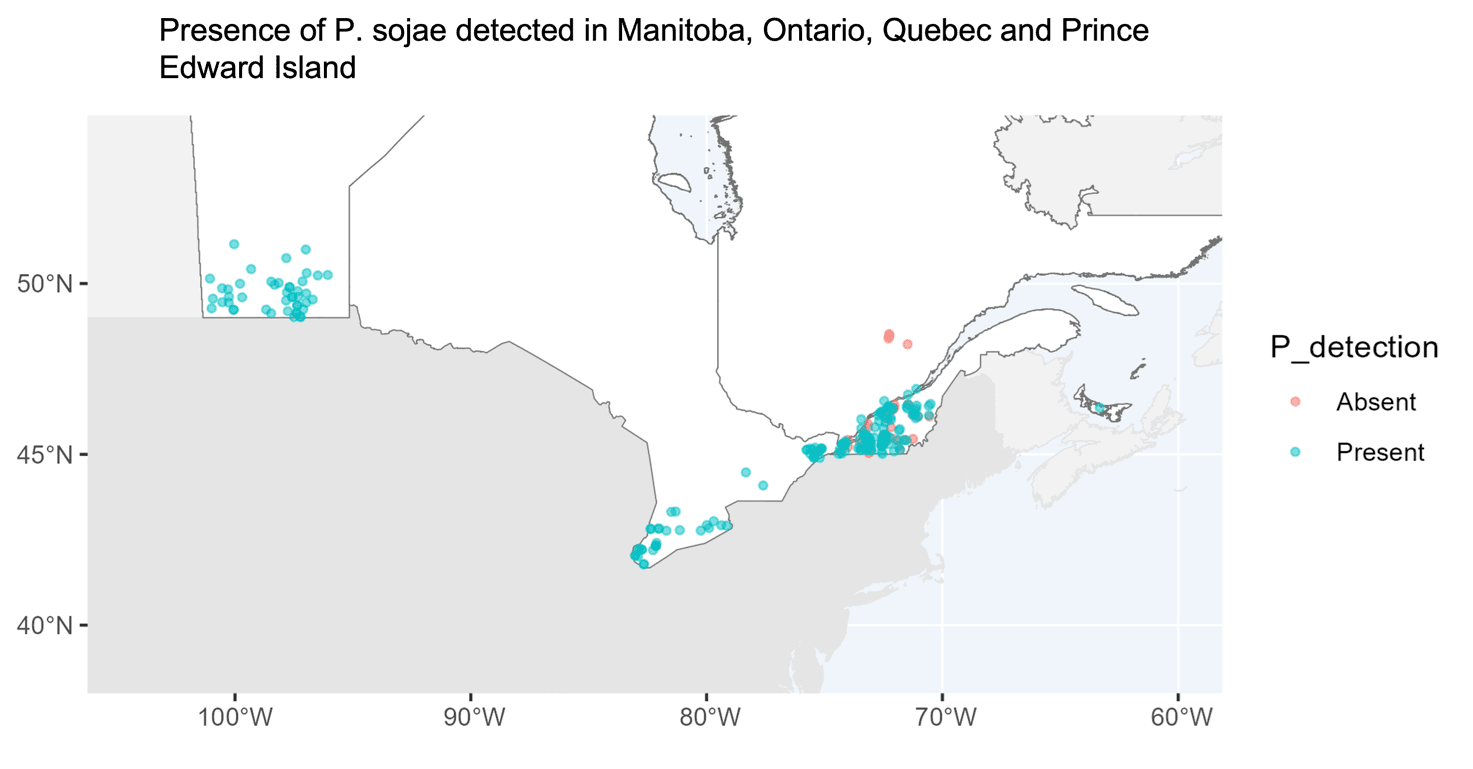
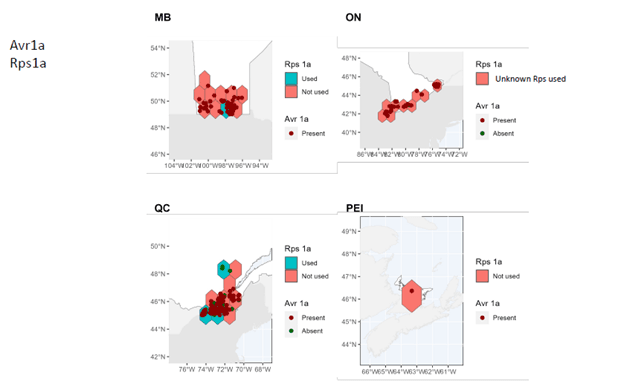
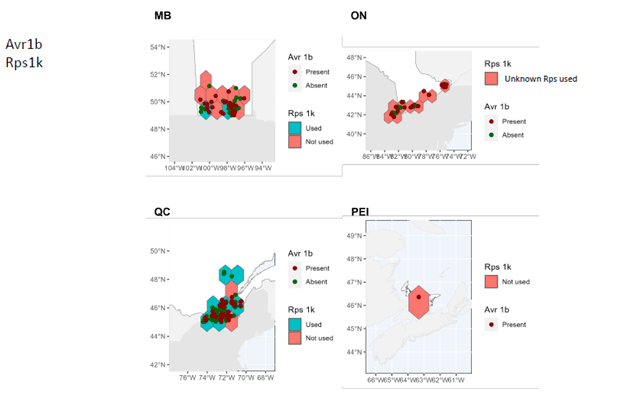
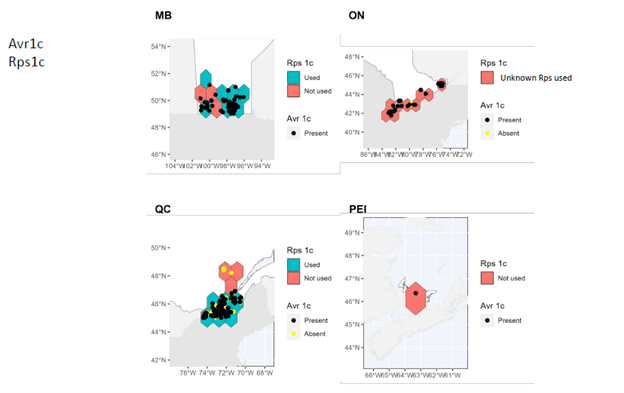
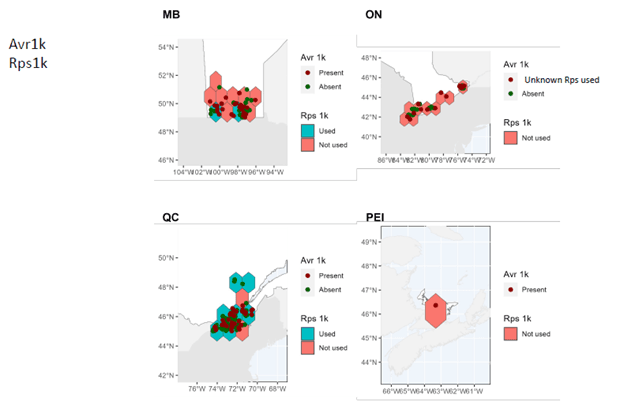
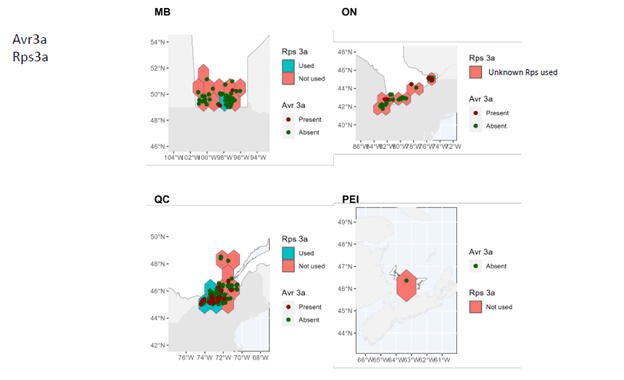
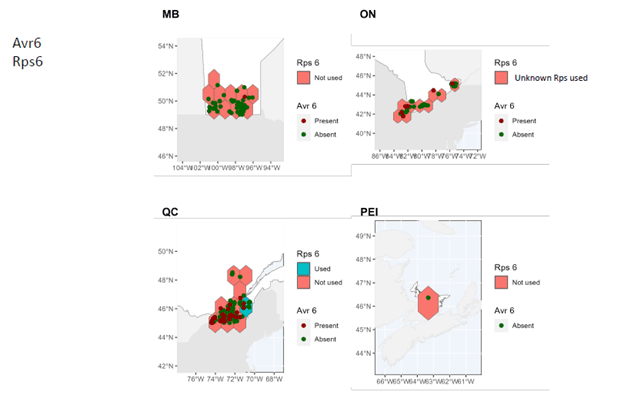
In Manitoba, Ontario, and Prince Edward Island, pathotypes 1a, 1b, 1k, 1c are very widespread while 3a and 6 are much less so. In Quebec, pathotypes 1a, 1c are very common. Although pathotypes 1b and 1k, 3a and 6 have been detected, several regions of Quebec do not yet have them.
It can therefore be seen that Rps3a and Rps6 are the resistance genes that could be effective in the majority of cases in the fight against P. sojae in all of the four provinces studied. It remains important to properly diagnose the pathotypes present before choosing a variety with the appropriate resistance genes in order to limit the selection pressure exerted by the resistance genes. Systematic use of the same resistance genes can rapidly lead to the development of resistance in strains of P. sojae, which drastically decreases the long-term effectiveness of the Rps genes.
These maps also highlight the importance of continuing to identify and introduce new Rps genes into commercial soybean lines to enable long-lasting genetic control.
Yield experience based on the right Rps gene
After each year of sampling, the identified pathotypes of P. sojae were provided to growers and agronomists. Soybean producers participating in this project applied the recommendations made. Based on the pathotype identified in their field, a disease-resistant soybean variety was planted the following year. This simple choice of the right gene resulted in a yield increase up to 40%. In collaboration with the Prime-Vert program (MAPAQ), a project was developed aimed at comparing the yields obtained among producers between four experimental plots in the same field. To do this, two plots were sown with a soybean variety possessing a resistant Rps gene and two others with a very similar variety except for an Rps gene susceptible to the disease (Figure 1). Photographed using a drone, the experimental plots likely had several areas of mortality or lacked vegetation cover. At harvest, the plots were weighed separately and those sown with the resistant variety experienced a weight yield gain of almost 50%.

Figure 1: Comparison of yields of soybean plots in 2022 planted with a variety containing the right Rps gene (resistant) versus a variety susceptible to the P.sojae variants present in the field. Yields in the resistant plots were 49% higher.
External Funding Partners:
This activity was funded in part by the Government of Canada under the Canadian Agricultural Partnership’s AgriScience Program, with industry support from the Canadian Field Crop Research Alliance (CFCRA) whose members include: Atlantic Grains Council; Producteurs de grains du Quebec; Grain Farmers of Ontario; Manitoba Corn Growers Association; Manitoba Pulse & Soybean Growers; Saskatchewan Pulse Growers; Prairie Oat Growers Association; SeCan; and FP Genetics.
Project Related Publications:
Belzile, F., Jean, M., Torkamaneh, D., Tardivel, A., Lemay, M. A., Boudhrioua, C., Arsenault-Labrecque, G., Dussault-Benoit, C., Lebreton, A., de Ronne, M., Tremblay, V., Labbé, C., and Bélanger, R. 2022. The SoyaGen Project: Putting genomics to work for soybean breeders. Frontiers in Plant Science. 1092.
McCoy, Belanger R.R. et al. 2023. A global-temporal meta-analysis perspective on Phytophthora sojae resistance-gene efficacy for disease management. Nature Communications.
Santhanam, P., Labbé, C., Tremblay, V., Bélanger, R.R. 2024. A rapid molecular diagnostic tool to discriminate alleles of avirulence genes and haplotypes of Phytophthora sojae using high-resolution melting analysis. Mol Plant Pathol. Jan: 25(1).
Vanessa, T., Debra, L., McLaren, Y.M.K., Strelkov, S.E., Conner, R.L., Wally, O., and Bélanger, R.R. 2021. Molecular assessment of pathotype diversity of Phytophthora sojae in Canada highlights declining sources of resistance in soybean. Plant Disease. 105 (12): 4006-4013.

